Clémentine Fillinger, Stéphanie Dumont, Patrick Vuillez, Etienne Challet

Presentation
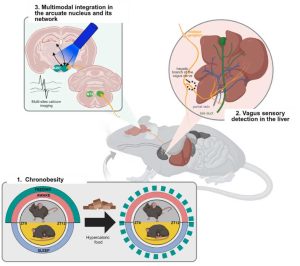 Junk food can affect the circadian organization of food consumption, leading to a pathological phenomenon that we named “chronobesity”. The arcuate nucleus of the hypothalamus (ARC) is a major food clock in the brain, located at the bottom of the third ventricle and close to the median eminence. The ARC is a critical sensing gateway for circulating nutrients and hormones and contains two main cell types: the orexigenic agouti-related peptide/neuropeptide Y-coexpressing neurons (AgRP/NPY), which signal hunger and promote food intake, and the pro-opiomelanocortin-expressing neurons (POMC), which selectively inhibit food intake and reduce body weight gain. At a neuroanatomical level, the ARC can be depicted as an integrating hub, receiving light-related signals from the suprachiasmatic nuclei (SCN), the brain’s light-entrainable master clock, with food-related information from peripheral organs relayed by the vagus nerve in the nucleus of the solitary tract of the medulla (NTS). The ARC also interacts strongly with other brain regions known to regulate feeding behavior and appetite, but the organization and synchronization of this complex network in chronobesity remains largely unknown. In the laboratory, we are studying the ARC and its connections with the body and the brain in male and female mice in response to hypercaloric food-induced dysregulation of feeding rhythm. Using cutting-edge approaches such as fiber photometry recording, DREADD, tissue clearing and rodent behavior, we aim to understand how this complex body-to-brain network can be perturbed by a simple change in diet, with a particular focus on the involvement of the hepatic component of sensory peripheral afferences.
Junk food can affect the circadian organization of food consumption, leading to a pathological phenomenon that we named “chronobesity”. The arcuate nucleus of the hypothalamus (ARC) is a major food clock in the brain, located at the bottom of the third ventricle and close to the median eminence. The ARC is a critical sensing gateway for circulating nutrients and hormones and contains two main cell types: the orexigenic agouti-related peptide/neuropeptide Y-coexpressing neurons (AgRP/NPY), which signal hunger and promote food intake, and the pro-opiomelanocortin-expressing neurons (POMC), which selectively inhibit food intake and reduce body weight gain. At a neuroanatomical level, the ARC can be depicted as an integrating hub, receiving light-related signals from the suprachiasmatic nuclei (SCN), the brain’s light-entrainable master clock, with food-related information from peripheral organs relayed by the vagus nerve in the nucleus of the solitary tract of the medulla (NTS). The ARC also interacts strongly with other brain regions known to regulate feeding behavior and appetite, but the organization and synchronization of this complex network in chronobesity remains largely unknown. In the laboratory, we are studying the ARC and its connections with the body and the brain in male and female mice in response to hypercaloric food-induced dysregulation of feeding rhythm. Using cutting-edge approaches such as fiber photometry recording, DREADD, tissue clearing and rodent behavior, we aim to understand how this complex body-to-brain network can be perturbed by a simple change in diet, with a particular focus on the involvement of the hepatic component of sensory peripheral afferences.
In parallel, to detect differential responses of the SCN and the two food-entrainable clocks (ARC and cerebellum) to nutritional synchronisers, we will study in vitro the chronomodulator effects of lipids and sugars on organotypic explants from PER2::Luc mice. Afterwards, we will phenotype the SCN, ARC and cerebellar cells affected using a Luminoview.

Clémentine Fillinger

Stéphanie Dumont

Patrick Vuillez